Contents
SCALEPACK outputs an ASCII (human-readable) "flat" file with the reflection data:
1
-985
108.547 160.025 270.755 90.000 90.000 90.000 p212121
0 0 6 -5.1 10.1
0 0 10 3034.3 153.9
0 0 12 840.6 49.0
0 0 14 651.1 43.0
0 0 16 2475.6 136.3
0 0 18 8179.2 388.2
0 0 20 29222.3 1326.0
I have no idea about the first two cryptic lines but the third line
contains your unit cell dimensions and space group. If you've got the
REFERENCE FILM 1line in your scale.com/scale.in file - or you've requested Global Refinement in HKL2000 - then the cell dimensions are the post-refined (i.e. optimal) ones out of Scalepack. The remaining lines in the file contain reflection data. For data output without the SCALE ANOMALOUS or ANOMALOUS flags, the reflection data all looks like:
0 0 6 -5.1 10.1 0 0 10 3034.3 153.9 0 0 12 840.6 49.0 0 0 14 651.1 43.0 0 0 16 2475.6 136.3 0 0 18 8179.2 388.2 0 0 20 29222.3 1326.0which is h, k, l, intensity, sigma(intensity) : the Miller indices of the reflection, it's estimated averaged intensity and the standard deviation thereof.
For data output with the anomalous flags, it looks more like:
.
.
.
1 0 38 17916.5 524.3
1 0 39 6971.1 225.9
1 0 40 11223.0 344.4
1 0 41 1858.6 119.9
1 0 42 180.7 78.3
1 0 43 132.9 84.1
1 1 1 814.6 22.2
1 1 2 22016.1 667.7
1 1 4 0 -1 5599.1 148.8
1 1 5 0 -1 16369.3 331.3
1 1 10 74860.2 2886.0 69208.2 1818.2
1 1 11 24627.8 818.8 30576.5 941.0
1 1 12 9907.5 398.1 10845.7 403.4
1 1 13 5758.6 250.4 5909.7 256.7
1 1 14 412.8 53.4 486.1 53.8
.
.
.
Centric reflections contain only h,k,l,Int,sigma(Int) because
they have no anomalous difference. Acentric reflections should
have Int(h,k,l) and Int(-h,-k,-l) - [we'll call this I(+) and I(-)
from now on] on the same line with the format
h,k,l,I(+),sigma(I(+)),I(-),sigma(I(-)) . For reflections with I(+)
missing the values are replaced with 0 and -1 - see the (1,1,4) and
(1,1,5) reflections in the above example which have I(-) observed but
are missing I(+) observations. For reflections with I(-) missing the
reflection just looks like a Centric reflection (e.g. the (1,1,1) and
(1,1,2) reflections in the above example) with just the I(+) values
listed. If you're not clear what a Centric and Acentric reflection
is, then Centric reflections have phase values restricted one of a few
values (typically 0 and 180 degrees). Centrics usually have one
Miller index of zero, but this doesn't guarantee that they are Centric
- e.g. there are none in the P1 space group. For a formal definition
of what makes a Centric reflection see: CCP4's maths
for protein crystallographers on this very subject.
Scalepack format gotcha: in what seems like a dubious design decision, those people at HKL make Scalepack change format for really strong reflections that do not fit within the standard format:
.
.
.
8 1 5 167487 3612
8 2 2 168299 6854 178745 6459
8 2 3 133771 2903 125313 3261
8 3 3 144493 3074 136694 5072
8 3 5 164070 3787 142771 4140
.
.
.
i.e. the decimal point is removed, allowing 100x larger numbers to be stored
in the same column width.
All this makes it very ugly to start writing your own data conversion program - leave it to the pros.

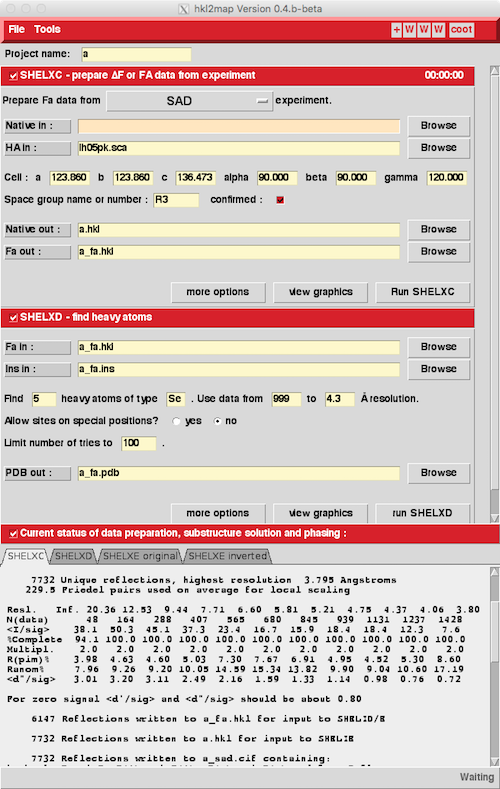
You can sequentially run SHELXC and SHELXD and then run SHELXE with
the two possible absolute structures. To view the phasing solutions
in e.g. Coot you'll have to convert the .phs SHELXE phase files into
MTZ format - these have a naming scheme like a_m20_s.5.phs and
a_m20_s.5_i.phs - the latter for the inverted enantiomorph. HKL2MAP
writes a script called something like a_phs2mtz.csh to convert the phs
files into mtz format. Sometimes the script needs a little tweaking
(e.g. change spacegroup from R3 to H3 if using the hexagonal setting)
but it's just using CCP4's F2MTZ and CAD programs to do the
conversion.
SHELXC uses Scalepack files directly so there's no conversion process - but SHELXC does expect the Scalepack files to have a .sca extension to you might have to do some renaming (I personally always use .sca as a Scalepack output file extension).
A typical SHELXC/SHELXD/SHELXE script looks like:
#!/bin/csh -f # # Script to run SHELXC/SHELXD to find heavy atoms in the MAD case # # SHELXC prepares Fa coefficients for SHELXD to chew on, finding the # effective resolution of the MAD/SAD/SIRAS data by correlation between # wavelengths (where possible) or signal/noise ratios. # It reads SCALEPACK files, WHICH MUST HAVE A .sca EXTENSION and contain # anomalous differences (use the ANOMALOUS keyword in Scalepack). # (if you have a low-energy remote it's keyword LREM, if you have a # reasonably isomorphous native you can include it with keyword NAT) # # (Files with extension .hkl are assumed to be SHELX files, which will # give the wrong result if they are really scalepack files) # # Change: names of files, cell dimensions, spacegroup, and number of sites # to FIND. The number of tries can be left at 50 and then sharply reduced # if SHELXD obviously finds a solution quickly. # # The name "se1" and "se1_fa" are the job names. The .lst # (log), .res (result) files are all given that name with various extensions # ## ## SHELXC prepares the data - the instruction (.ins) file is given the job ## name with _fa appended to it. ## # /usr/local/shelx/macosx/shelxc se1 << EOF HREM se1rm.sca PEAK se1pk.sca INFL se1in.sca LREM se1lo.sca CELL 35.104 63.496 76.458 90. 90. 90. SPAG P212121 FIND 8 NTRY 50 EOF # # ## ## Actually run SHELXD - the best result is in the .res file, the log file ## is in the .lst file, results also listed to the screen ## # /usr/local/shelx/macosx/shelxd se1_fa # ## ## Run solvent flattening in SHELXE with 35% solvent for 100 cycles ## Include the -h flag if you don't have NATIVE data included in ## SHELXC (since your "native" data for flattening will contain the heavy atom) ## The -i flag tries the inverse of the heavy atom solution ## Supposedly the one with the highest contrast is the correct one. Note that ## inverting the sites will also invert the space group (e.g. P3<1>21 goes ## to P3<2>21) ## ## The program automatically picks up the best result from the .res file ## produced by SHELXD ## # /usr/local/shelx/macosx/shelxe se1 se1_fa -s0.45 -m60 /usr/local/shelx/macosx/shelxe se1 se1_fa -s0.45 -m60 -i #After SHELXC the program(s) use their own reflection format with the extension .hkl (consult the documentation). SHELXC is a pretty good program for analyzing your MAD data while you are collecting it (d''/sig signal/noise ratio, for example).
#!/bin/csh -f # ## ## Scalepack2MTZ takes SCALEPACK output files (with or without anomalous ## data) and converts them to MTZ files containing the same intensity data ## For anomalous data it also computes Imean and SIGImean ## # scalepack2mtz HKLIN bxc-063.sca HKLOUT bxc063.mtz << EOF ANOMALOUS NO SYMMETRY P212121 TITLE Convert scalepack data to MTZ format SCALE 10.0 EOF # ## ## Convert the INTENSITIES to STRUCTURE FACTORS ## If TRUNCATE NO is used the conversion is a simple SQRT() ## If TRUNCATE YES is used for estimating very weak reflections, which may ## have observed small negative I's, will be given small positive F's based ## on the expectation of the distribution derived from Bayesian statistics ## ## NRESIDUE <#residues> will attempt to put the data on an ABSOLUTE SCALE ## assuming you have an educated guess as to the a/u contents ## Absolute scales can be useful for analysis of difference maps ## ## Part of the truncate output gives a Wilson-like description of the ## project atomic/solvent content based on NRESIDUE ## # truncate hklin bxc063.mtz hklout bxc063_truncate.mtz << EOF TITLE Truncate run on native data TRUNCATE YES LABIN IMEAN=IMEAN SIGIMEAN=SIGIMEAN NRESIDUE 340 EOF # uniqueify bxc063_truncate.mtz # # mtz2various HKLIN bxc063_truncate-unique.mtz HKLOUT bxc063.free << EOF RESOLUTION 500.0 2.9 OUTPUT CNS FREEVAL 1 MISS 0.0 LABIN FP=F SIGFP=SIGF FREE=FreeR_flag END EOFIn SCALEPACK2MTZ we can override the cell dimensions, if you prefer, and define the space group which might be different to the arbitrary one you assigned during scaling. We tell if if there is anomalous data present or not using the ANOMALOUS keyword. For reasons of precision it's usually better to multiply the supplied data by a factor of 10. From the Scalepack .sca file, we get an MTZ-format file bcx063.mtz that contains essentially the same contents as the original .sca file. TRUNCATE converts intensities to structure factors, and puts the data on an absolute scale if you supply the number of residues in the asymmetric unit. Here we tell TRUNCATE to actually remap the intensity data (TRUNCATE YES) based on Bayesian statistics which can be useful for weak data where a decent chunk of the reflections would have |F|=0 which may then get omitted from CNS refinement. The uniqueify line adds randomly-selected FreeR flags to the MTZ file, creating bxc063_truncate-unique.mtz. We can then write the contents of this file out to CNS format using MTZ2VARIOUS for use in refinement.
This is a very simple script and many variants are possible. You can use the program MTZDUMP to probe the data stored in the binary (not human-readable) MTZ files.
Some attention needs to be paid to the output of TRUNCATE - in particular it gives you an estimate for the solvent content given the number of residues that you specified. This is useful when estimating the number per asymmetric unit. Also the cumulative intensity statistics can be a fairly good indicator of the potential presence of twinning (which is guaranteed to ruin your day).
If you put your MTZ file into X-PLOR format (like CNS format but without the headers) then you can use Todd Yeates' twinning server to test for twinning. Alternatively take a look at the Xtriage section below.
#!/bin/csh -f
#
##
## Scalepack2MTZ takes SCALEPACK output files (with or without anomalous
## data) and converts them to MTZ files containing the same intensity data
## For anomalous data it also computes Imean and SIGImean
##
#
scalepack2mtz HKLIN se1in.sca HKLOUT inflection.mtz << EOF
ANOMALOUS YES
CELL 35.104 63.496 76.458 90. 90. 90.
SYMMETRY P212121
TITLE Convert scalepack data to MTZ format
SCALE 10.0
EOF
#
scalepack2mtz HKLIN se1pk.sca HKLOUT peak.mtz << EOF
ANOMALOUS YES
CELL 35.104 63.496 76.458 90. 90. 90.
SYMMETRY P212121
TITLE Convert scalepack data to MTZ format
SCALE 10.0
EOF
#
scalepack2mtz HKLIN se1rm.sca HKLOUT remote.mtz << EOF
ANOMALOUS YES
CELL 35.104 63.496 76.458 90. 90. 90.
SYMMETRY P212121
TITLE Convert scalepack data to MTZ format
SCALE 10.0
EOF
#
scalepack2mtz HKLIN se1lo.sca HKLOUT lowremote.mtz << EOF
ANOMALOUS YES
CELL 35.104 63.496 76.458 90. 90. 90.
SYMMETRY P212121
TITLE Convert scalepack data to MTZ format
SCALE 10.0
EOF
#
scalepack2mtz HKLIN nat1.sca HKLOUT native.mtz << EOF
ANOMALOUS NO
CELL 34.859 63.271 76.360 90. 90. 90.
SYMMETRY P212121
TITLE Convert scalepack data to MTZ format
SCALE 10.0
EOF
#
##
## Convert the INTENSITIES to STRUCTURE FACTORS
## If TRUNCATE NO is used the conversion is a simple SQRT()
## If TRUNCATE YES is used for estimating very weak reflections, which may
## have observed small negative I's, will be given small positive F's based
## on the expectation of the distribution derived from Bayesian statistics
##
## Probably the simplest option is TRUNCATE NO, reserving TRUNCATE YES
## for difficult cases or if you want to fantasize about dropping your
## R-free by 0.1%
##
## NRESIDUE <#residues> will attempt to put the data on an ABSOLUTE SCALE
## assuming you have an educated guess as to the a/u contents
## Absolute scales can be useful for analysis of difference maps
##
## Part of the truncate output gives a Wilson-like description of the
## project atomic/solvent content based on NRESIDUE
##
#
truncate hklin inflection.mtz hklout inflection_truncate.mtz << EOF
TITLE Truncate run on SAD data
TRUNCATE NO
LABIN IMEAN=IMEAN SIGIMEAN=SIGIMEAN I(+)=I(+) SIGI(+)=SIGI(+) -
I(-)=I(-) SIGI(-)=SIGI(-)
NRESIDUE 180
EOF
#
truncate hklin peak.mtz hklout peak_truncate.mtz << EOF
TITLE Truncate run on SAD data
TRUNCATE NO
LABIN IMEAN=IMEAN SIGIMEAN=SIGIMEAN I(+)=I(+) SIGI(+)=SIGI(+) -
I(-)=I(-) SIGI(-)=SIGI(-)
NRESIDUE 180
EOF
#
truncate hklin remote.mtz hklout remote_truncate.mtz << EOF
TITLE Truncate run on SAD data
TRUNCATE NO
LABIN IMEAN=IMEAN SIGIMEAN=SIGIMEAN I(+)=I(+) SIGI(+)=SIGI(+) -
I(-)=I(-) SIGI(-)=SIGI(-)
NRESIDUE 180
EOF
#
truncate hklin lowremote.mtz hklout lowremote_truncate.mtz << EOF
TITLE Truncate run on SAD data
TRUNCATE NO
LABIN IMEAN=IMEAN SIGIMEAN=SIGIMEAN I(+)=I(+) SIGI(+)=SIGI(+) -
I(-)=I(-) SIGI(-)=SIGI(-)
NRESIDUE 180
EOF
#
truncate hklin native.mtz hklout native_truncate.mtz << EOF
TITLE Truncate run on native data
TRUNCATE NO
LABIN IMEAN=IMEAN SIGIMEAN=SIGIMEAN
NRESIDUE 180
EOF
#
#####
# MERGE all the MTZ files into one huge file
#####
#
cad HKLIN1 inflection_truncate.mtz \
HKLIN2 peak_truncate.mtz \
HKLIN3 remote_truncate.mtz \
HKLIN4 lowremote_truncate.mtz \
HKLIN5 native_truncate.mtz \
HKLOUT l123_dano.mtz << EOF
CELL 35.104 63.496 76.458 90. 90. 90.
SYMM P212121
RESOLUTION OVERALL 100000.0 1.0
TITLE Combined native and derivative data
LABIN FILE 1 E1=F E2=SIGF E3=DANO E4=SIGDANO
CTYP FILE 1 E1=F E2=Q E3=D E4=Q
LABOUT FILE 1 E1=F1 E2=SIGF1 E3=ANO1 E4=SIGANO1
LABIN FILE 2 E1=F E2=SIGF E3=DANO E4=SIGDANO
CTYP FILE 2 E1=F E2=Q E3=D E4=Q
LABOUT FILE 2 E1=F2 E2=SIGF2 E3=ANO2 E4=SIGANO2
LABIN FILE 3 E1=F E2=SIGF E3=DANO E4=SIGDANO
CTYP FILE 3 E1=F E2=Q E3=D E4=Q
LABOUT FILE 3 E1=F3 E2=SIGF3 E3=ANO3 E4=SIGANO3
LABIN FILE 4 E1=F E2=SIGF E3=DANO E4=SIGDANO
CTYP FILE 4 E1=F E2=Q E3=D E4=Q
LABOUT FILE 4 E1=F4 E2=SIGF4 E3=ANO4 E4=SIGANO4
LABIN FILE 5 E1=F E2=SIGF
CTYP FILE 5 E1=F E2=Q
LABOUT FILE 5 E1=FN E2=SIGFN
END
EOF
#
Here we've got FIVE runs of SCALEPACK2MTZ, one for the high energy remote,
peak, inflection and (unusually) the low energy remote and native data. We
flag all the MAD datasets with ANOMALOUS YES in SCALEPACK2MTZ. Conversion
is via TRUNCATE in each case, this time there are more labels to deal with
because of the presence of I(+) and I(-). Finally the CAD run is new: there
are five input files going into one output file. Each dataset has a
unique "F" column label (F1, F2, F3, F4, FN) and the MAD datasets have the
corresponding ΔAno column labels (ANO1, ANO2, ANO3, ANO4) plus the
standard deviations of all of those. The use of the RESOLUTION keyword makes
sure that all the input data makes it to the output. Note that because the
native and MAD datasets have different unit cells, we force a specific unit
cell within CAD for the purposes of eliminating inconsistency.
phenix.xtriage residues=240 my_data.scaproduces the desired behavior (defining the number of residues is rather useful). The twinning analysis is pretty good, and it also looks for the effects of ice rings and pseudo-centering to warn you of potential pitfalls in your data. You can use the L-test and other statistical tests to look for weird intensity distributions in the data that suggest twinning or pseudo-centering, both of which can have devastating effects on the ability to solve your structure. Xtriage is now more widely installed than previously, so most synchrotron beamlines should have it.
#!/bin/csh -f # # # mtz2various HKLIN bxc063_truncate-unique.mtz HKLOUT bxc063.cif << EOF RESOLUTION 50.0 2.5 OUTPUT CIF data_sf FREEVAL 1 MISS 0.0 LABIN FP=F SIGFP=SIGF FREE=FreeR_flag END EOF #and in fact this small script looks a lot like the mtz2various incarnation used to produce the CNS format reflection files. In fact it should be as identical as possible to avoid causing inconsistencies.